The following blog post was co-authored by Emily Watkins
Speed up core Unix operations like listing and searching datasets by 10x to 100x.
Listing Files Slows Down AI Workflows
Directory tree “walks” tend to be one of the major bottlenecks in deep learning workflows–sometimes without developers even realizing it.
For example, deep learning training jobs often begin by shuffling the training dataset. Shuffling enables the model to be trained iteratively using random subsets of training data. For shuffling to work, the application often must first list all file paths in a dataset, and this is often done with “os.walk”.
Like the Unix “ls” command, Python’s “os.walk” interface issues file system calls to sequentially traverse a directory tree. For datasets containing tens to hundreds of millions of files, sequential directory tree traversal can add anywhere from 10 minutes to hours of overhead before training even begins. Developers often want initial feedback about the model after the first couple batches of training (e.g. to see how they need to tune hyperparameters). So, long-running directory tree traversals at the start of every job can significantly slow down overall productivity.
To address the bottleneck of slow directory walks, it helps to first understand why listing file metadata has not scaled well as data sets have grown into the tens of millions of files.
Why Listing Large Datasets is a Challenge
AI workflows that shuffle datasets using Python’s filesystem libraries have similar bottlenecks to core Unix utilities–namely, that they list files sequentially rather than concurrently. That works fine when an application’s file system lookups return almost instantly, which is what happens when metadata requests result in cache hits for the local OS’s directory cache.
As datasets grow into the tens of millions of files, however, it becomes difficult for all of a file system’s metadata to fit in the local OS’s directory cache. When file metadata requests can no longer be serviced by the local OS’s cache, response times jump from the nanoseconds to microseconds and even milliseconds. A latency close to a millisecond for file system calls becomes very significant when sequentially listing millions of files.
For example, let’s imagine that an application can list a directory’s contents in half a millisecond. That means, if it doesn’t utilize concurrency, this application can list around 2,000 directories per second, or around 120,000 directories per minute. If each directory contains, say, 20 files on average, the application can list 2 million files a minute, or 150 million files in an hour. That means a billion file dataset would take all day to list one time.
Solutions to address metadata performance challenges often include additional infrastructure. To support efficient querying of metadata, customers utilize metadata caches, sophisticated file formats, special software, or non-standard kernel modules designed to manage, cache, and shuffle file metadata efficiently. This leaves customers with an unpleasant tradeoff between the performance and complexity of their data pipeline.
RapidFile Toolkit = Faster Unix Commands
Together, the Pure Storage® RapidFile Toolkit and FlashBlade™ address the performance vs. complexity tradeoff by using familiar command line interfaces to provide high-performance, multi-threaded Linux utilities optimized for FlashBlade’s highly-concurrent NFS storage.
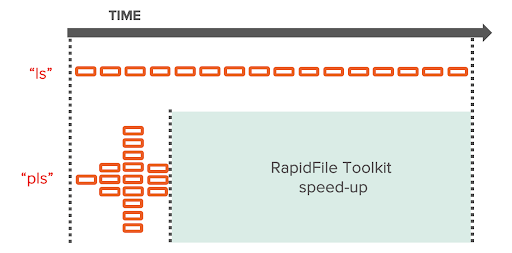
RapidFile Toolkit’s “pls” and “pfind” preserve the familiar command line interfaces of “ls” and “find” but utilize a userspace NFS client library, libnfs, to issue multiple concurrent RPCs for listing file system metadata, bypassing Linux core utilities’ decades-old serial codepaths.
As sub-directories are discovered, RPCs to walk them are added to a queue and executed in parallel.
When listing file metadata of massive data sets stored on FlashBlade, we’ve seen speedups between 10x (on low latency networks) and 100x (on higher latency networks) when switching from standard Unix commands or Python’s os.walk to the RapidFile Toolkit.
Shuffling a Dataset with the RapidFile Toolkit
Here we present a shell script that uses RapidFile Toolkit’s pfind, reservoir sampling, and the Fisher-Yates shuffle to generate a random subset of 1000 file paths out of a larger directory structure containing 5 million files.
In our tests on FlashBlade, this script finished in 33 seconds.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 |
COMPLETE_DATASET=/mnt/flashblade/5MFiles SHUFFLED_DATASET_SIZE=1000 pfind “$COMPLETE_DATASET” –type f | \ awk –v SIZE=“$SHUFFLED_DATASET_SIZE” \ ‘BEGIN { srand() } !/^$/ { \ if (NR <= SIZE) { set[NR–1]=$0; } \ else { r=rand(); j=int(r*(NR–1)); if (j<SIZE) set[j]=$0; } \ } END { \ for (i = SIZE – 1; i > 0; i—) { \ j = int(rand() * (i + 1)); \ swap = set[i]; \ set[i] = set[j]; \ set[j] = swap; \ } \ for (i=0; i<SIZE; i++) {print set[i];} } |
Integrating RapidFile Toolkit into Jupyter notebooks
In one of our lab environments, we host Jupyter as a service for our data scientists. It enables easy, automated onboarding for new users, and it provides a centralized way to manage our Jupyter configuration.
In this blog post, we demonstrate providing the RapidFile Toolkit by default in user environments by adding it to the related Docker image. This Docker image could be used either in a JupyterHub environment (for something like Jupyter-as-a-Service) or as a one-off by a single developer locally.
Obtaining the RapidFile Toolkit
Pure Storage FlashBlade customers can download the toolkit using their Pure1® login.
If you’re not a customer, reach out to info@purestorage.com to set up a demo.
Installing RapidFile Toolkit into a Docker Image
In this example, we assume you have already downloaded the toolkit as a file named “purestorage-rapidfile-toolkit.tar”.
Add the following section to the Dockerfile that you will be using for your data science environment. Commands in this example are based on a Debian/Ubuntu base image. Adding this code to the tail end of the file should be fine.
|
1 2 3 4 5 |
USER root ARG RAPIDFILE_VER=1.0.0–beta.4 ADD purestorage–rapidfile–toolkit.tar /purestorage–rapidfile–toolkit.tar RUN tar –xvf /purestorage–rapidfile–toolkit.tar \ && dpkg –i rapidfile–$RAPIDFILE_VER/rapidfile–$RAPIDFILE_VER–Linux.deb \ |
Once you deploy the Docker image as the basis for a Jupyterhub environment, “pls” and other commands in the RapidFile Toolkit are now available automatically for data scientists.
Example Usage
We have several large training datasets saved in our shared NFS storage, which is mounted to the directory “/home/shared/”.
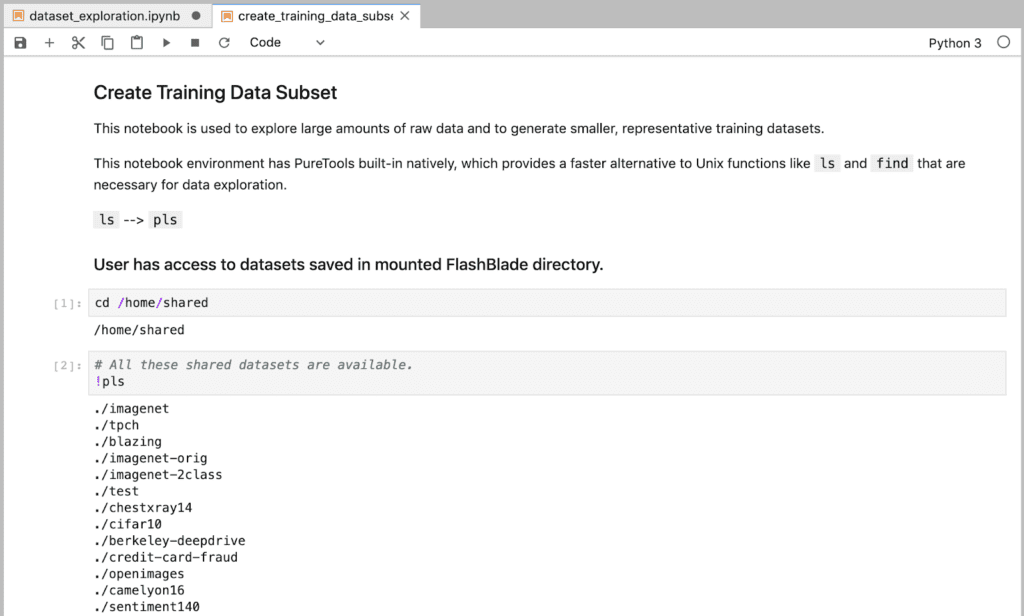
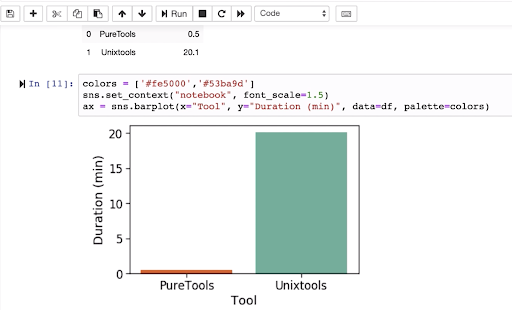
With one of our example training datasets, we’re able to list its contents 40x faster.
Conclusion
RapidFile Toolkit combined with Pure Storage FlashBlade can dramatically speed up AI workflows without complex infrastructure by removing the bottleneck for listing file metadata.
It’s easy to get the performance advantages of RapidFile Toolkit on FlashBlade in a range of other workloads as well, such as by integrating “pls” into Spark, or in EDA simulations. Stay tuned for further blog posts about RapidFile Toolkit.