This post is the first of a multi-part series by Bikash Choudhury and Emily Watkins, where we will discuss how to configure a Kubernetes-based AI Data Hub for data scientists.
Recently, my son, a freshman studying Computer Science in college, asked me “Why Python for Data Science? It’s slow.” He had just started his first AI project during an on-campus hackathon event. Lucky for him, he was using Jupyter notebooks (ipynb) in Google cloud (Colaboratory). Environments like it bundle the full stack of components necessary to deliver a fast python environment. It’s far more than Python alone.
In this blog post, I’ll describe why it’s worthwhile to build your own on-premise data science platform. With an AI Data Hub in place, you can host Jupyter notebooks and kickstart data scientists’ projects–and also start to automate more of the other applications that are part of your machine learning pipelines. Because you have performant storage backing the hot tier of datasets used to build deep learning models, you can put that storage to use hosting persistent volumes for all the other aspects of the AI Data Hub.
Software for an AI Data Hub
As data scientists explore data sets, they might clean, transform, or run training and inference on them. Jupyter notebooks are a popular Interactive Desktop Environment (IDE) for data science work in Python.
To get to the goal of hosting Jupyter notebooks for data scientists, we have to examine a wide array of applications.
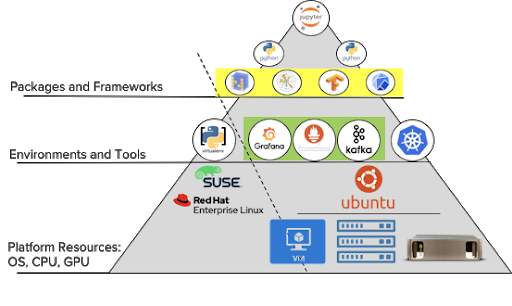
We’ll review the configuration from the user downwards, starting with Python as the coding language.
Planning for Self-Service
As my son experienced with his Google-hosted Jupyter notebooks, it is possible to give data scientists a quick, self-service place to start coding. It requires slightly more setup and planning from the infrastructure team, but it leads to greater efficiency overall. Workspaces for new users to the platform (or additional compute or storage needs of one user) can be provisioned automatically, saving time for both the data scientist and the IT team.
Why Kubernetes?
Kubernetes uses Docker images to package not just the Python libraries but also dependencies like applications, tools, and optional OS packages. It’s easy for data scientists to deploy their various Docker images on Kubernetes. Multiple containers with the same environment can be deployed on a single node – or multiple nodes – depending on a workload’s CPU and GPU requirements.
Advantages of using Kubernetes to host data scientists’ Python environments are:
- Applications are run as microservices in containers that can scale and are easy to debug.
- Kubernetes manages the CPU and memory resources efficiently when the user either logs out or shuts down the Jupyter server.
- Declarative configuration with YAML allows the reproducibility of experiments across pipelines and platforms.
- The Kubernetes ecosystem is growing at a rapid pace with various new tools data scientists can use in their ML pipelines.
While Kubernetes provides a layer for application virtualization, it runs on the host OS. Of all the popular variants of Linux, Ubuntu has been a common choice of platform in the AI/ML workflow.
- Ubuntu natively supports Python libraries like Mathplotlib, TensorFlow, and NumPy as well as NVIDIA’s CUDA drivers for GPUs.
- Ubuntu has a strong library and framework ecosystem with great community support.
- Ubuntu provides a standard platform in hybrid cloud for application (container) portability in Kubernetes clusters on-premise or in the public cloud.
Our example AI Data Hub described in the following posts is based on Ubuntu, but other deployments could follow the same solution stack using another OS.
Choosing the Infrastructure for the Environment
After maneuvering through the different layers top-down, the success of the AI platform depends on access to data.
Ideally, teams should move or stream AI data sources to a single data platform like Pure Storage FlashBlade™. FlashBlade functions as a Data Hub to store and process data through the various phases of the AI/ML workflows.
Data moves between the different stages of AI pipelines from the time raw data is ingested, cleaned, and transformed to train and test models, deployment, and inference. However, it’s much simpler–and faster–if the data doesn’t have to actually physically move. With a performant storage server that all the compute nodes can access, the datasets themselves don’t have to actually move.
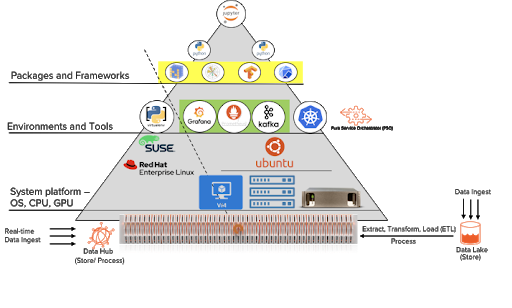
Traditionally, the Extract, Transfer, Load (ETL) process can be time and resource-intensive. Storing and processing data directly on FlashBlade provides better efficiency, performance, and manageability.
Acting as a data hub, FlashBlade provides persistent storage for many AI workflows in a Kubernetes cluster. FlashBlade can store everything from Jupyter notebooks themselves to model checkpoints, to monitoring data from the Kubernetes cluster. In fact, in an upcoming blog post, we’ll describe how to optimize ancillary applications like Docker registries and version control repositories with FlashBlade.
Conclusion
We’ve described the base that sets the stage for an AI Data Hub where Data Scientists can start their experiments. On such a data hub, Jupyter notebooks can use persistent storage from FlashBlade to save and share test results among team members. Jupyter-as-a-Service allows data scientists to on-board and start their exploratory work quickly.
The data hub can also host the various applications that continuously scale as data scientists evolve their workflows, like Kafka, and Spark.
Stay tuned next week for the next post in this multi-part series: Hosting Jupyter-as-a service on FlashBlade. We’ll cover the configuration steps needed to host Jupyter-as-a-service backed by FlashBlade. Read further posts in the series over the coming weeks:
- Storing a Private Docker Registry on FlashBlade S3
- Scraping FlashBlade metrics using a Prometheus exporter
- Visualizing Prometheus data with Grafana dashboard for FlashBlade
- Automating an inference pipeline in a Kubernetes Cluster
- Tuning networking configuration of a Kubernetes-based AI Data Hub
- Integrating Pure RapidFile Toolkit into Jupyter notebooks